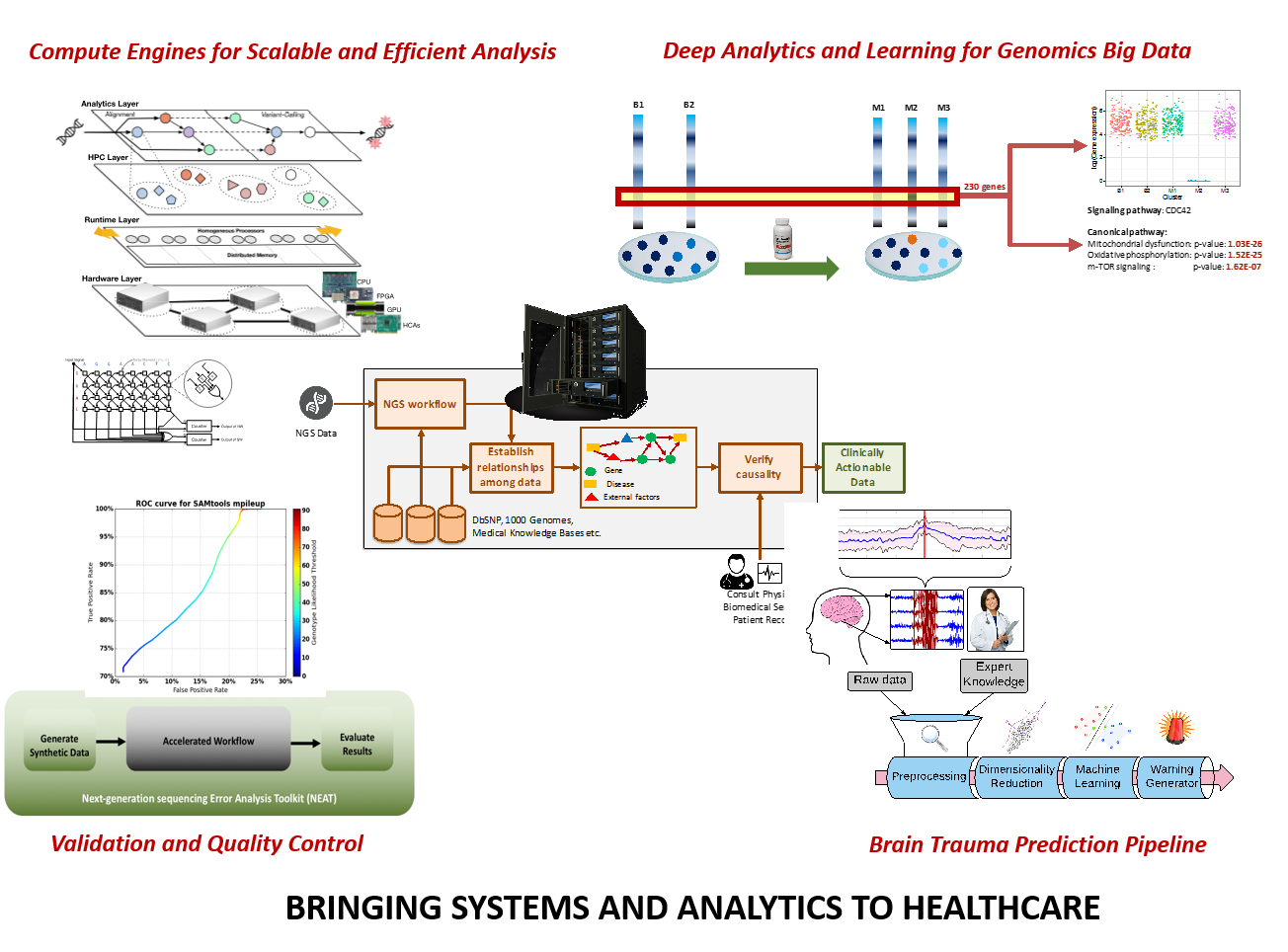
Individualized or precision medicine is a big idea on the horizon of biomedical research and practice. Key to the success of individualized medicine is the integrative analysis of clinical and biological big data, such as genomics, metabolomics, with longitudinal data, such as individual patient health records. These integrated and deep analytics can lead to small but significant insights useful in making crucial decisions of diagnosis and targeted treatment. Such insights are referred to as actionable intelligence. A major challenge in achieving individualized medicine is to efficiently provide actionable intelligence to physicians, enabling them to make clinical decisions that are more refined, accurate, and effective, especially in more complex and rare cases.
Developing a framework with the integrated analytics is nontrivial, requiring not only rigorous engineering fundamentals, but also close partnering with clinical and biological experts. The DEPEND group at the University of Illinois works closely with the Mayo Clinic, the National University of Singapore, and a select group of computing, pharmaceutical, and biotechnology companies to address the challenges of achieving actionable intelligence. The DEPEND group has been engaged in advanced data analytics and engineering systems research to address a range of difficult medical problems using as inputs a combination of big data, patient-specific medical records, clinical trials, and physician records. The value, generality, and specificity of their methods can be seen in the breadth of applications being developed and the extent of the group’s collaborations with physicians, biologists, and bio-informaticians. Some examples of the diseases addressed in the current work include cancer pharmacogenomics, depression, neurological and degenerative brain diseases including seizures and Alzheimer’s, and chronic diseases like diabetes.
To advance this work, we have brought to bear a range of advanced real-time analytics and specialized machine-learning methods, including supervised and unsupervised learning, probabilistic graphical models, and real-time analytics. The group has deployed a variety of computational resources, including high-performance computing at NCSA, advanced cloud computing, and specialized FPGA and GPU-based computing engines and special-purpose accelerators.
Health Analytics
In individualized medicine, working closely with Drs. Liewei Wang, Richard Weinshilboum, Rani Kalari, Mark Frye and William Bobo at Mayo Clinic’s Center for Individualized Medicine, we have developed tools and technologies i) to use supervised machine learning methods to predict drug responses in depression patients treated with antidepressants using metabolomics, genomics, and clinical data, ii) to use model-based unsupervised machine learning methods to infer drug response in triple-negative breast cancer using single-cell RNASeq, and iii) to predict drug response in triple-negative breast cancer patients using game theoretic models and RNASeq data. This work has resulted in two major awards from both the machine learning big data community as well as the pharmacogenomics community.In collaboration with surgeons and endocrinologists at Singapore’s National University Hospital using advanced graphical models (factor graphs), we have developed the SingaDragn framework to predict and preempt surgical readmissions and long-term health complications from electronic health records of diabetic patients in Singapore.
Health Systems
CompGen
Broader Impact
Illustrative of the breadth our work, Arjun Athreya’s collaboration with various colleagues has resulted in significant contributions to: 1) understanding metformin’s mechanism in treating triple-negative breast cancer (with Drs. Weinshilboum, Wang, and Kalari at Mayo Clinic), 2) applying a game-theoretical approach to understanding the epigenetics of adenocarcinoma and preeclampsia, with results now being applied to breast cancer (with Prof. Derek Wildman and his research team at the Carl R. Woese Institute for Genomic Biology), and 3) predicting therapeutic outcomes in diabetes (with Dr. Kee Yuan Ngiam at the National University Hospital in Singapore). The overarching goal of the research is to combine patient-specific genomic biomarkers with a variety of patient data to predict clinical outcomes. Enabling individualized diagnoses requires extraction of actionable intelligence, such as accurate biomarkers, and then the ability to predict clinical outcomes using extracted biomarkers for each patient. These techniques enable physicians and surgeons to be more proactive. Multidisciplinary research of this kind, much neglected in the past, is now viewed by biologists, physicians, and computer scientists and engineers as crucial for progress in their respective fields.