Research Staff:
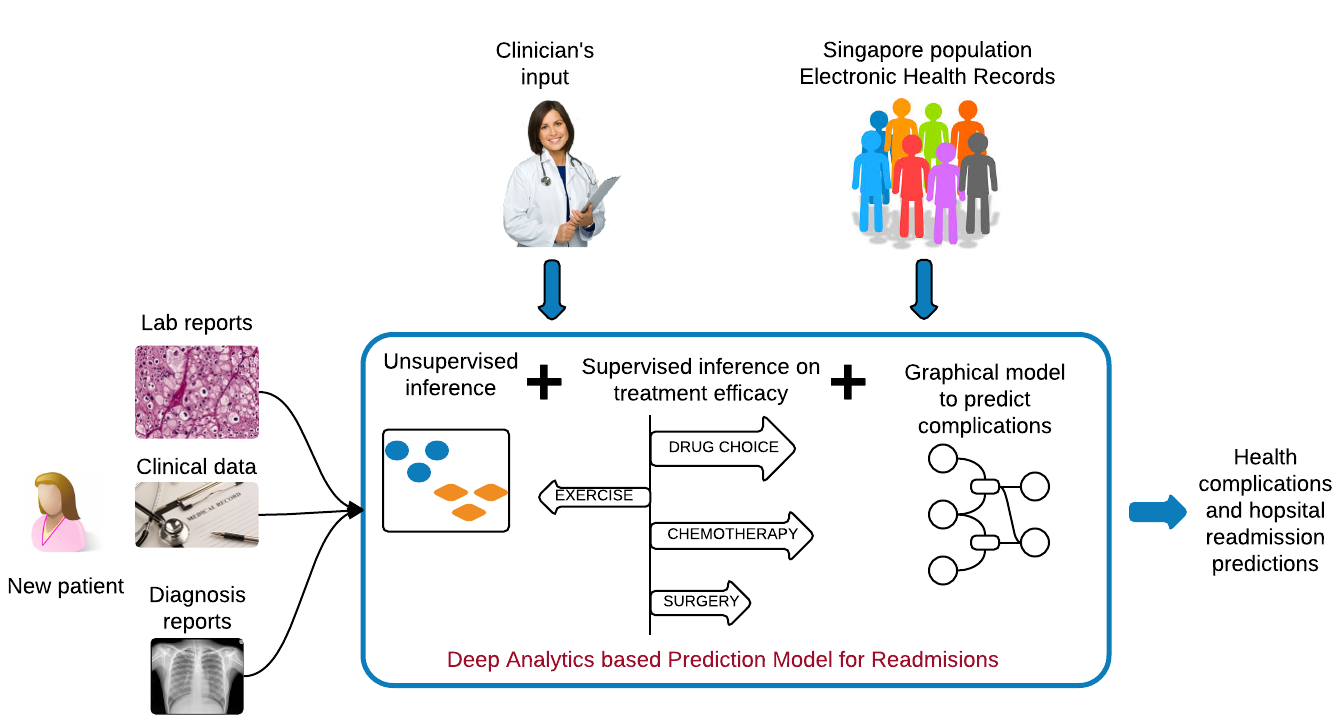
The overarching objective of the analytics in our multi-omic analyses as represented in each disease type will be to understand drug response mechanisms and to make patient-specific predictions for drug response; a pressing need under the Precision Medicine Initiative. Addressing this problem is key to Precision Medicine Initiative’s success because not all patients respond to a drug, and the drug response varies by the patient.
Identification of disease sub-types
This project identifies disease sub-types in depressed patients using a combination of unsupervised learning and probabilistic graphical models such as the hidden Markov model. The project, in collaboration with Drs. Liewei Wang and Richard Weinshilboum, Mayo Clinic uses clinical covariates, quantified symptoms of depression and metabolomics data from the largest single-site study at the Mayo Clinic.
Inferring mechanisms of drug response in disease subtypes
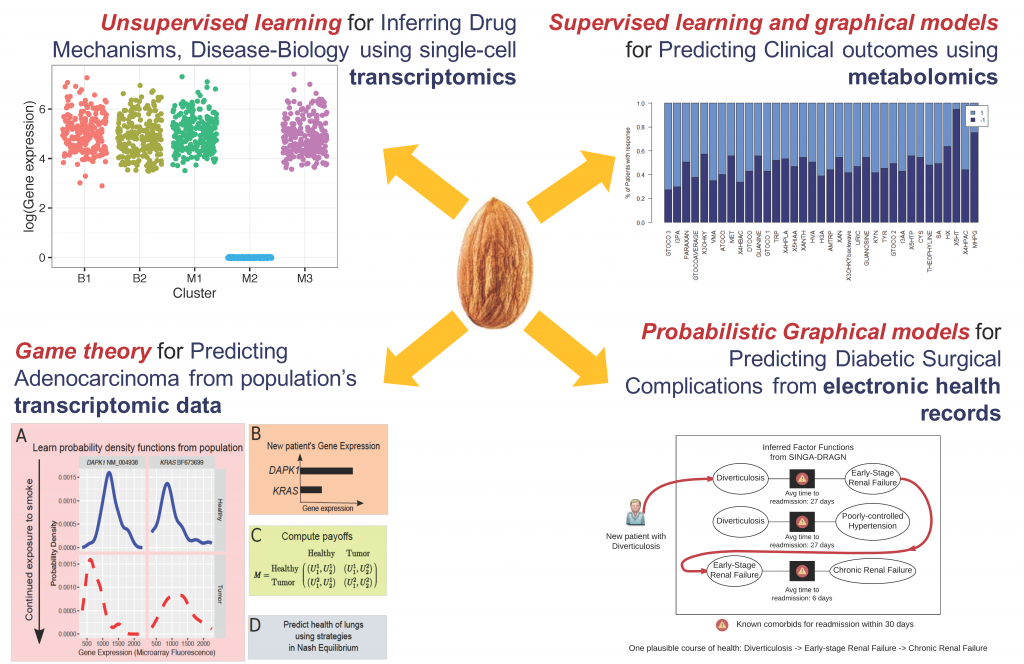
We infer mechanisms of drug response in disease sub-types, in the context of an anti-diabetic drug metformin and triple-negative breast cancer (TNBC), a molecular subtype of breast cancer for which there are no targeted therapeutics, yet. In collaboration with Drs. Junmei Cairns, Rani Kalari, Liewei Wang and Richard Weinshilboum, Mayo Clinic, we have developed a model-based unsupervised learning tool, Mixture Model-based Single-cell Analysis (MiMoSA) to infer cell-types induced by metformin in breast cancer cells. Combined with MiMoSA’s findings and pathway analysis, one particular gene, CDC42 was chosen for functional studies. Lab experiments verified that, down regulation of CDC42 by metformin inhibited cancer cell migration and proliferation in an AMPK-independent mechanism. This is a novel finding, as metformin’s actions are known to be AMPK-dependent.
Predicting risk of disease progression and drug response in lung adenocarcinoma and triple-negative breast cancer
Using competing relationships from the directionality in differentially expressed genes, we have built a game theoretic model to predict the likely phenotypes observed based on transcriptomic analyses. Our model has high accuracies in, a) evaluating the risk of lung adenocarcinoma from smoking (collaboration with Prof. Derek Wildman, Univ. Illinois) and, b) drug response in TNBC patients of the BEAUTY trial at Mayo Clinic (collaboration with Drs. Rani Kalari, Liewei Wang and Richard Weinshilboum, Mayo Clinic).
Predicting disease-related hospital readmission using electronic health records
It is important to understand population-wide risk and predispositions to certain health conditions. This allows for a cost-benefit model for early surgical interventions to prevent future complications, thereby aiding in graceful aging and reducing healthcare costs. In collaboration with Drs. Kee-Yuan Ngiam and C. N. Lee, National University Hospital, Singapore, we have analyzed the electronic health records of the diabetic population of Singapore by using Factor Graphs, a type of probabilistic graphical model, to predict long-term likely health complications of an individual and the expected timeline to such complications. Given the volume of records being analyzed, the tool we developed was run using Blue Waters, a supercomputing facility at the Univ. of Illinois.