Research Staff:
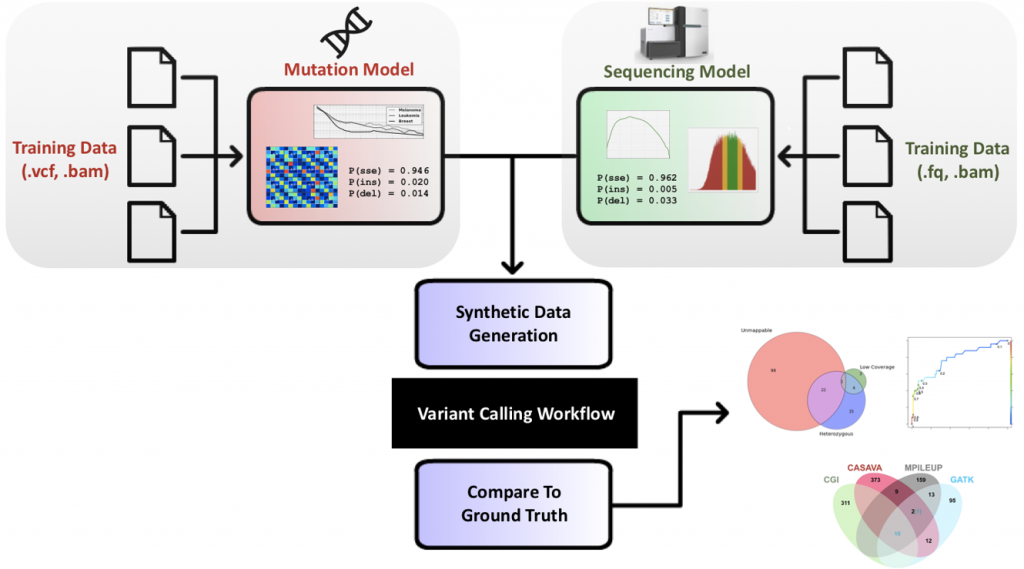
The benchmarking and validation of personalized medicine workflows is essential to ensure the integrity of clinical decisions that are increasingly incorporating genomic data. Our research in this area includes a simulation framework for benchmarking variant calling workflows, quantifying the impact of various error sources on workflow output, and developing new algorithms that leverage upcoming sequencing technologies. Our genomic data simulation tools are in use at multiple institutions, such as NCSA and the Ontario Institute for Cancer Research, and emulate many nuances of how mutations are distributed and how reads are sequenced based on real data. In ongoing work we are developing methods for detecting and visualizing arbitrarily complex structural variation from long read data. Our approach is showing unmatched accuracy for deciphering genomic rearrangements that cannot be confidently inferred using short reads.